Open Targets has entered a new phase: the Open Targets Genetics (OTG) portal is now fully integrated into the Open Targets Platform. Variant, study, and credible-set data now sit alongside target discovery in one interface. Researchers can interpret gene-disease evidence from both common and rare variation, across multiple ancestries, all without switching between tools. This consolidation reduces fragmentation, providing teams with a single, trusted resource for genetics-driven discovery.
In the March and June 2025 releases, Open Targets introduced three new entity types:
- Variant (~6.5M)
- Study, and
- Credible Set
They also rolled out a refreshed Locus-to-Gene (L2G) model. With SHapley Additive Explanations (SHAP), users can now see which features contribute to gene prioritisation, making results more transparent and interpretable. Colocalisation is anchored in credible set overlaps, creating a more consistent fine-mapping experience.
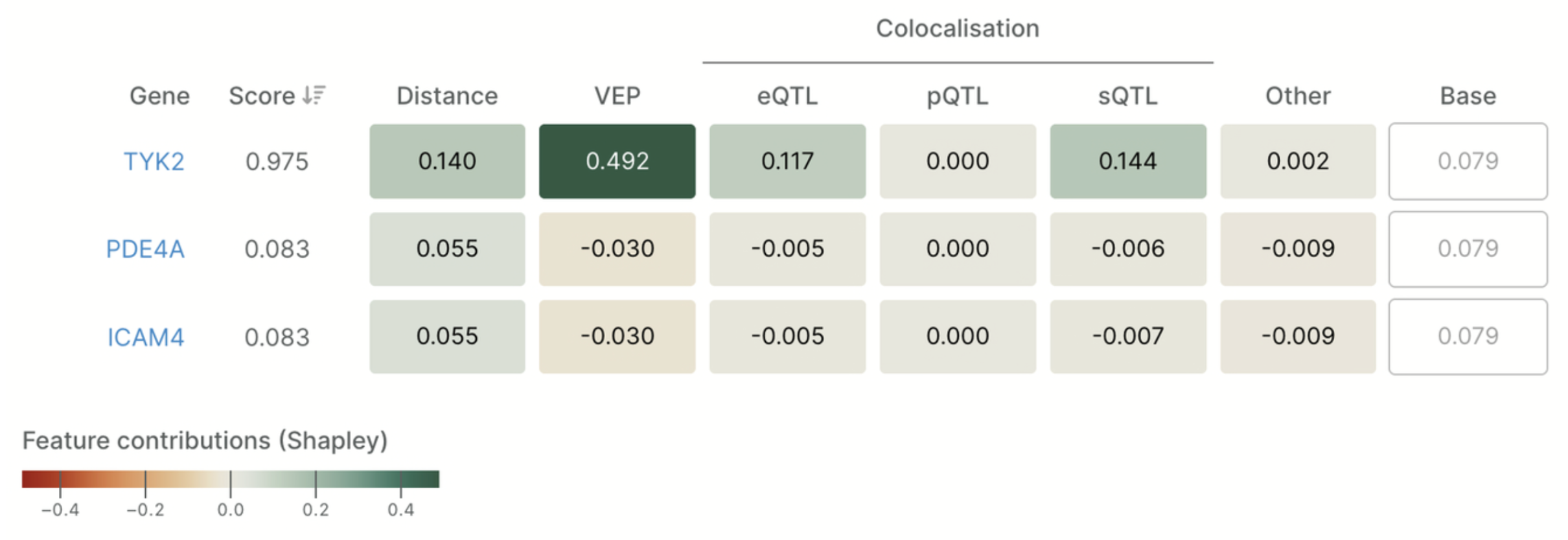
The L2G model utilizes SHAP (Shapley Additive Explanations) to quantify the contributions of various evidence types, including variant distance, QTLs, and functional annotations, to the prioritization of candidate genes. In this example, TYK2 receives the highest score for a credible set linked to psoriasis, supported mainly by VEP and eQTL features.
The June release (25.06) expanded coverage substantially, with 36% more GWAS credible sets (driven in part by the VA Million Veteran Program) and new burden evidence from the Broad CVDI Human Disease Portal. Open Targets Platform downloads now follow the Croissant metadata standard, making it easier to discover and reuse data programmatically.
On the UI side, you’ll notice a faster global search, pharmacogenetics directionality, and an updated molecular structure viewer (AlphaFold context with AlphaMissense pathogenicity). Additionally, Associations on the Fly provides richer filtering, pinning, and inline data inspection to facilitate quick triage of signals.
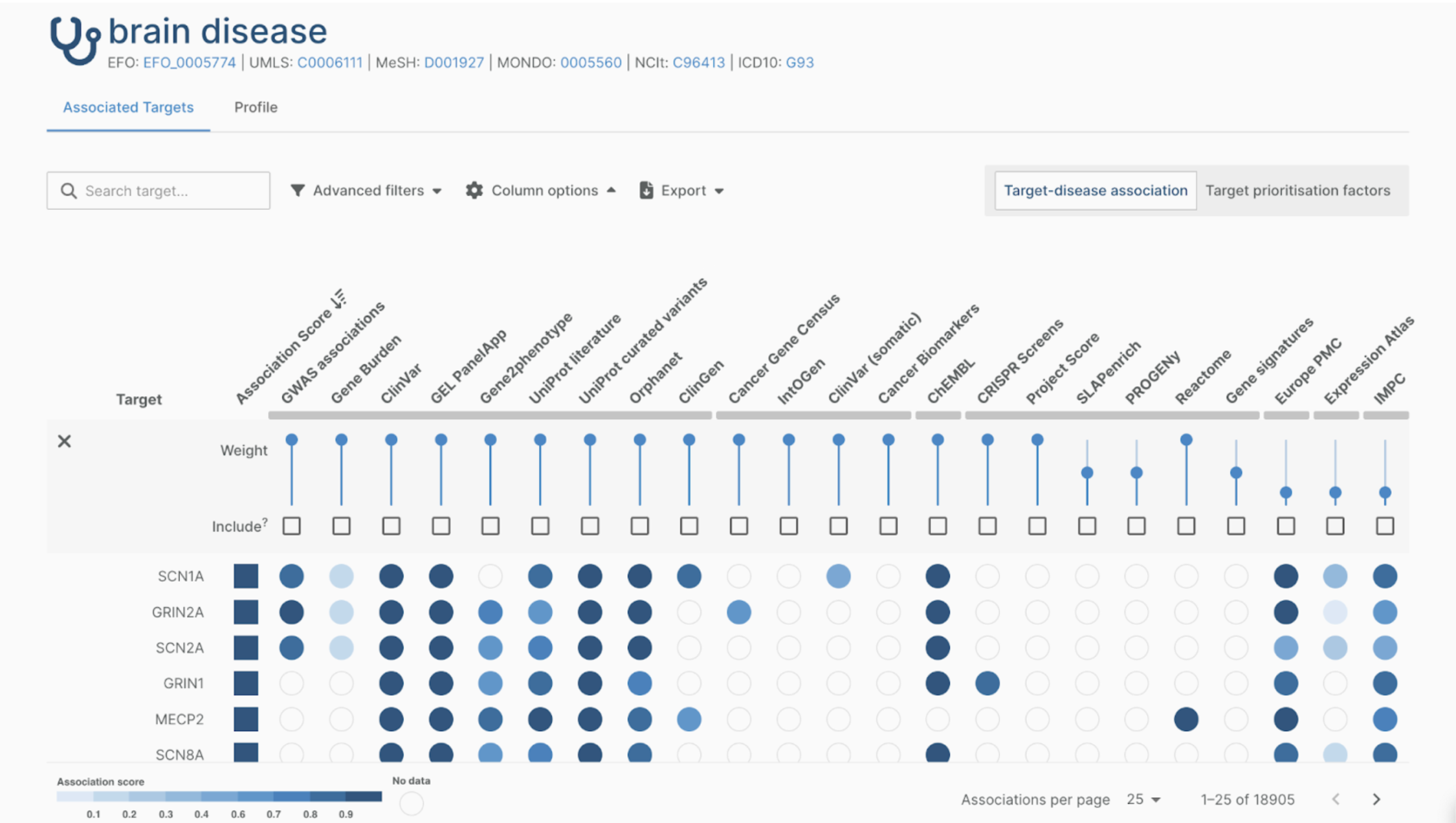
The new “Associations on the Fly” view allows users to filter, pin, and inspect evidence inline, making it easier to triage and prioritise gene–disease associations quickly.
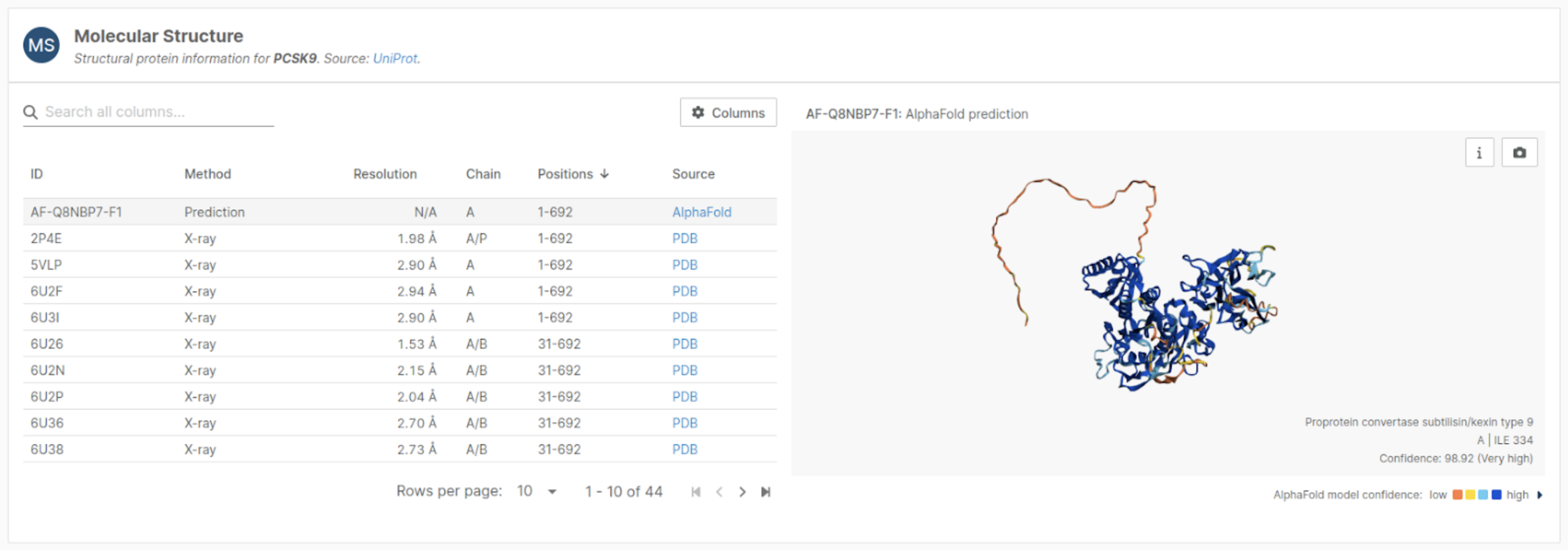
The updated molecular structure viewer combines AlphaFold predictions with experimental data, enriched with AlphaMissense annotations, to provide context on variant pathogenicity.
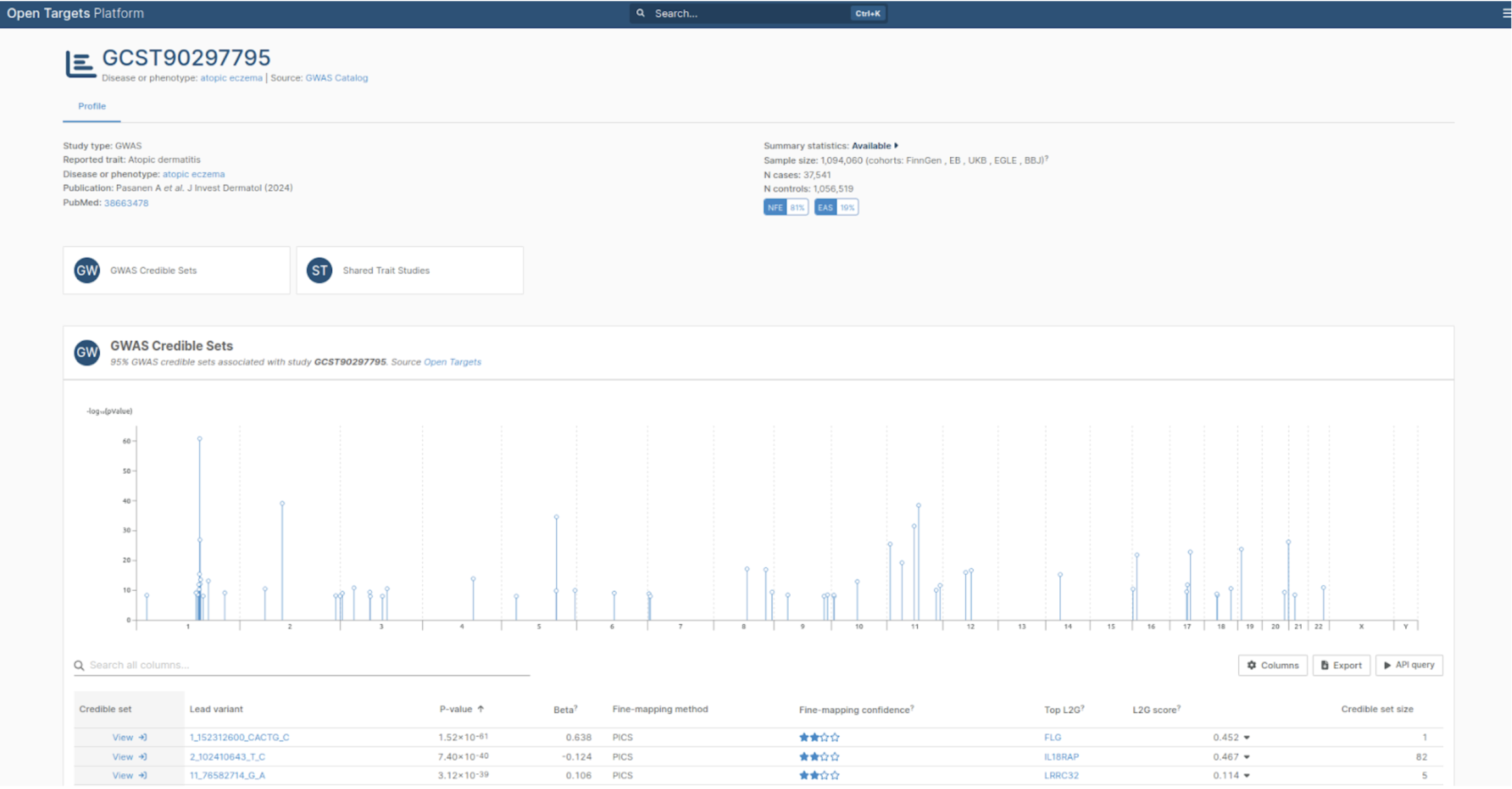
The Platform now displays GWAS study results with Manhattan plots and credible sets, helping researchers assess fine-mapping confidence and interpret genetic associations at a glance.
With the standalone Genetics UI deprecated in July 2025, organisations should plan for upgrade and consolidation paths to the unified Open Targets Platform.
How The Hyve supports your adoption
- Deploy & Upgrade: We install/upgrade the Open Targets Platform (cloud or on‑prem), configure secure auth, automate builds, and migrate users from the legacy Genetics UI.
- Integrate your data: Our engineers set up robust ETL pipelines to harmonise your internal genetics/omics data into Open Targets, aligned with Croissant metadata for reproducibility.
- Run Gentropy: We operationalize Gentropy for fine-mapping, colocalization, and L2G at scale, from notebook trials to orchestrated production runs.
- Enable your teams: Role‑based training for scientists and data teams, SOPs, dashboards, and SLAs aligned to Open Targets’ quarterly cadence.
If you’d like to see Open Targets in action, you can explore The Hyve’s demo server: platform.gtd.thehyve.nl. The demo illustrates how the Platform can be customised and extended with additional modules such as cBioPortal data integration and knowledge graphs.
This flexibility, combined with our deployment and training services, ensures that your organisation can adapt Open Targets to its specific genetic needs and target discovery workflows.
References: Open Targets release notes & docs; NAR 2025 Platform paper (Oxford Academic).