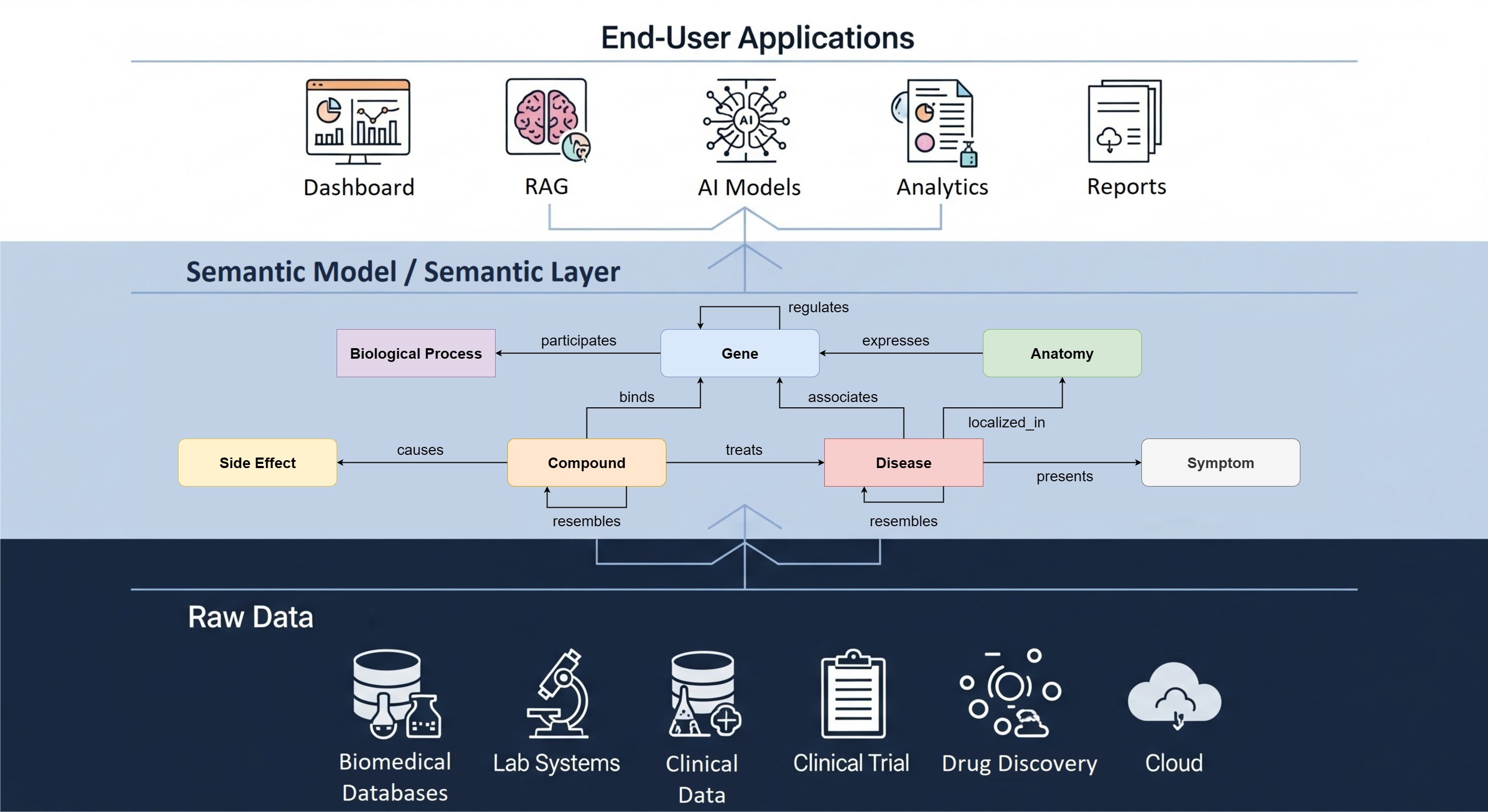
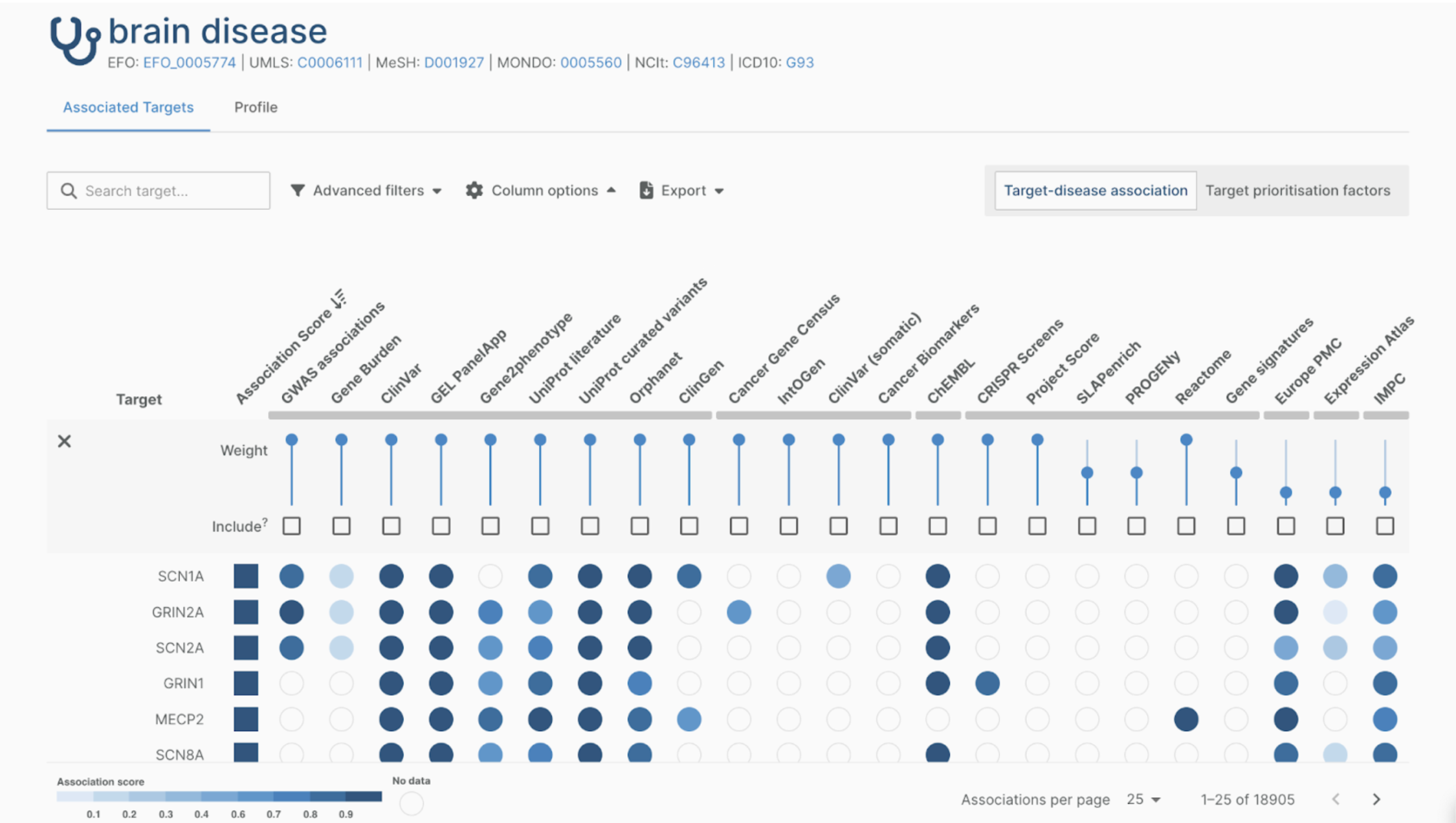
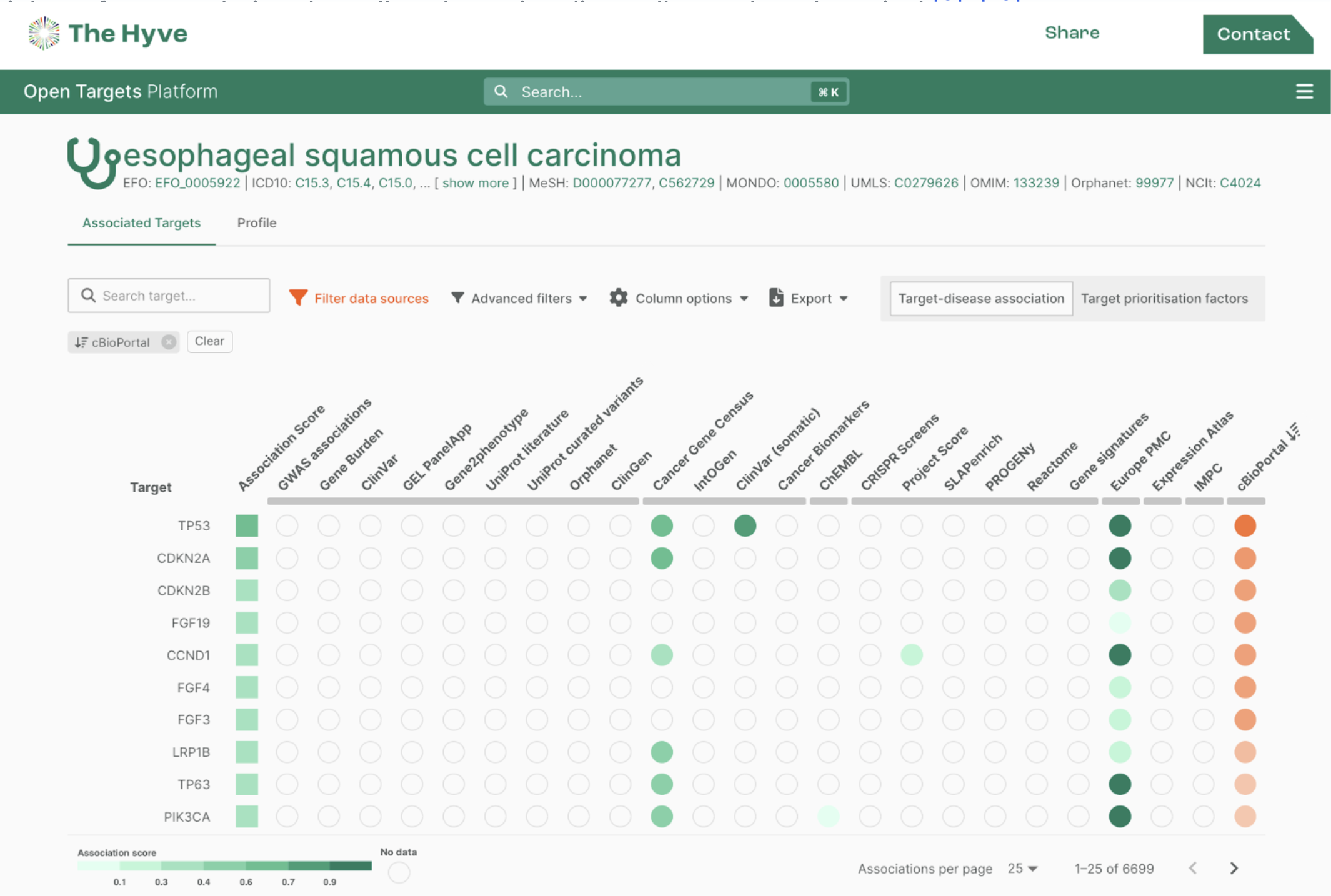
Battling standardization in metabolomics
BETHESDA, MD – Imagine a snow-covered Washington DC, just before Christmas. That was the scene where metabolomics researchers from around the world headed on December 15, 2010. They gathered at the National Institutes of Health to tackle a problem that has been on their radar for a few years: the standardization of reporting for metabolomics experiments. Granted, it’s an intrinsically complex task: differences in handling protocols, data acquisition, experimental designs and data processing all affect the experimental outcome, not to mention the effects of measurement and biological variation! And despite efforts, started already in 2005 by the Metabolomics Society, to organize the community and write up and implement those standards, the field still lacks the standardized repositories of compounds, spectra and experiments which would enable researchers to re-use each others’ data. However, the discussion is slowly moving from the content and procedures to the actual efforts and governance structures that need to be in place to really implement this standardization. If one thing became clear at this meeting, it is that the will and vision for pulling this off is present, but there are a few missing key pieces. For example, no publishers were present at the meeting. Also, actual collaborations on programming and database levels are still limited. And there is still a lot to develop on the level of standardization itself, within and outside the scope of this initiative: how to uniquely describe not only complete, but also partially known chemical structures, which ontologies to use for the description of biological samples, or how to organize the curation of uploaded chemical structures (let’s make a computer game out of it!) The workshop, organized by the Pharmacometabolomics Research Network, will have a number of follow up sessions in the coming year on dedicated topics, so stay tuned via http://metaboknow.org!