cBioPortal is widely known as a user-friendly tool for visualizing and analyzing cancer genomics data. cBioPortal is Open Source and can be installed locally, for instance to host patient data that is not allowed to leave the hospital firewall, or to analyze proprietary research data. A lesser known fact about cBioPortal is that it can be configured to work with mouse data as well. This blog post aims to highlight this underappreciated possibility.
Use cases
cBioPortal is used both for clinical decision making as well as cancer research. Although cBioPortal is primarily used for human data, there are a number of good reasons and use cases for use with mouse data.
Mouse model catalog
When starting an experiment to test a scientific hypothesis, it is very important to select the right models for the experiment. This decision can be based on both genomic or phenotypic data and depends on the goal of the experiment. The intuitive and, to most users, already familiar interface of cBioPortal can help both technical and non-technical users to find a model fit for the experiment with ease.
In addition to speed and convenience, there may be an even bigger advantage to having a mouse model catalog. In large organisations it can happen that a researcher is creating his own mouse model for his experiment because he is simply not aware of the existence of a model that is already suitable for the job. It could also be the case that an appropriate mouse model is already available at one of the contract research organizations (CRO). A catalog of all available models can prevent this issue. Obviously, this is also beneficial for the CROs, as they can allow their customers to search their mouse model catalog in an already familiar interface.
Translational research
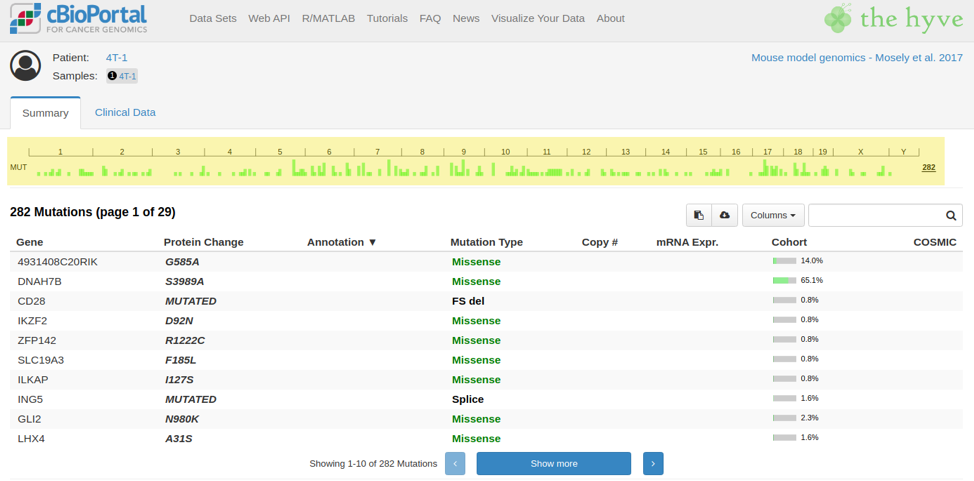
After completion of the mouse model experiments, data can be loaded into cBioPortal. Firstly, this increases the reusability (FAIRness) of this data, which makes it easier to use in future experiments. Secondly, cBioPortal serves as a great resource for generating new hypotheses and doing translational research in general. Since this is already a common use case for cBioPortal with human data, we will not go into the details in this blog post, as the use cases are quite similar.
How does it work?
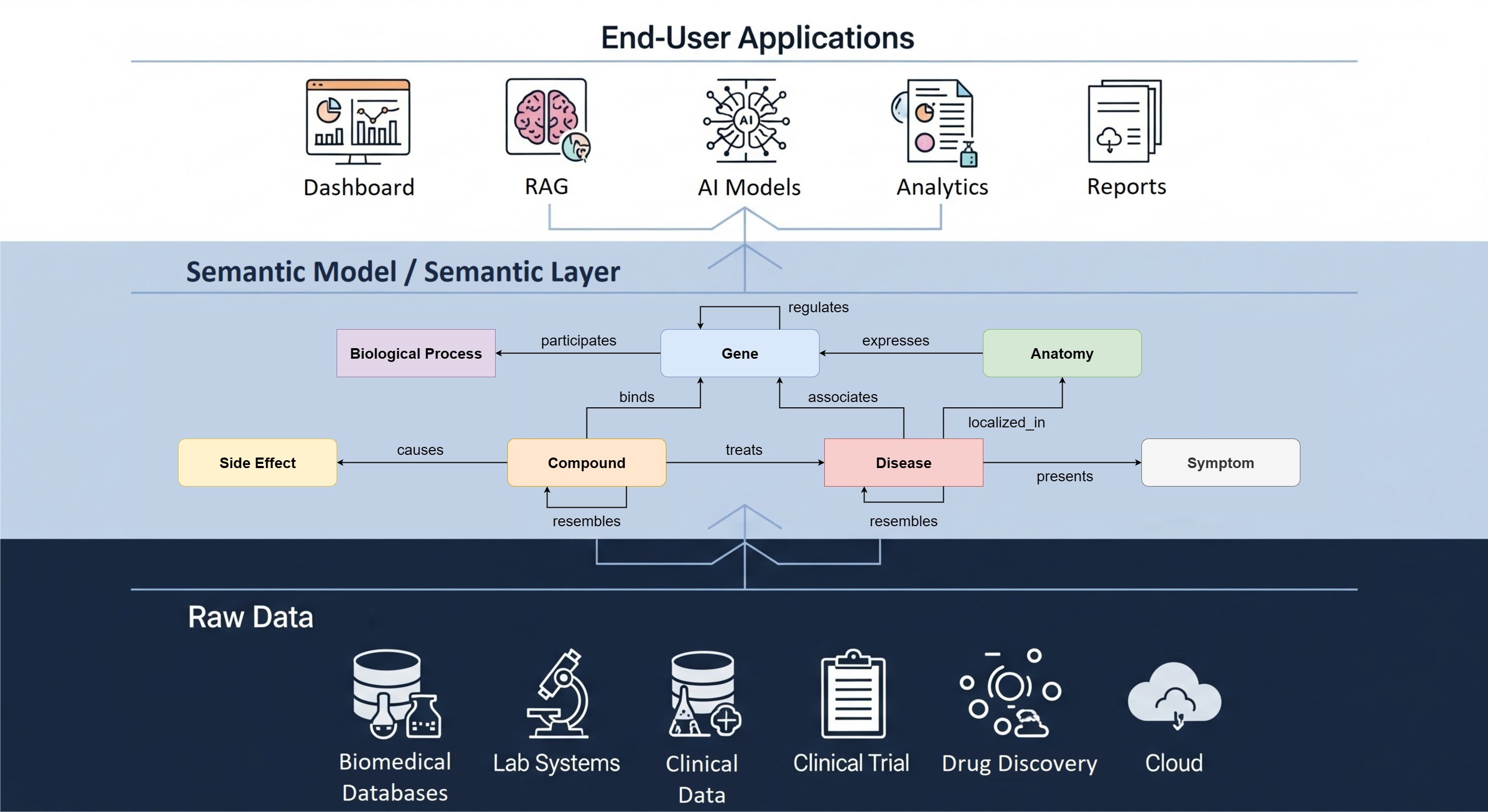
Large parts of cBioPortal are completely independent of the gene database and the reference genome used but there are a few exceptions.
One example is the set of gene names that a user can type into the query page and which show up in the interface on almost all views. Mouse genes often have the same name as their human orthologs, for instance for BRAF, but there are also genes where this is not the case, such as TRP53, which is known as TP53 in human. To remedy this, we replace the contents of the genes table by loading an alternative seed database containing mouse genes.

Another example is in the mutations view, where gene lengths and protein domains are used to display a lollipop plot. Mouse genes can have different lengths and protein domains from their human counterparts. This data is provided by Genome Nexus, which is a separate web service from cBioPortal that provides these types of background information. To have a fully functional cBioPortal for mouse, you would have to set up a local instance of Genome Nexus that is based on the mouse reference genome and gene databases. For convenience, we already included a mouse database in the Genome Nexus repository.
In the ‘patient’ view there is a genome overview that shows the number and length of the chromosomes. A setting in the portal properties needs to be changed to select the GRCm38 build. This bit is still hard coded and adding a new potential species to cBioPortal would require some code changes (but not many).
Closing remarks
Multiple genome builds
You may have heard that cBioPortal supports multiple genomes, or that this is being worked on. This is correct to the extent that it is possible to have multiple genomes in the backend. However, since it is not yet possible to have cBioPortal talk with multiple instances of Genome Nexus, the gene lengths could be incorrect, resulting in odd looking visualizations in the front end. We advise our clients to set up a separate instance for mouse, in particular because it also does not make much sense to mix human and mouse samples in analyses.
Other species
Mouse models are by far the most used animal models in cancer research but there are other animal models that are used for specific types of cancer as well, for instance dogs or pigs. The successful deployment of a cBioPortal instance for mouse paves the way to include other animals as well. Note that for patient-derived xenograft (PDX) models, cBioPortal for human can be used, since they are made up of human tissue.
What we do
The Hyve provides services to develop, extend and improve features in cBioPortal. This includes setting up cBioPortal for mouse and loading mouse data into it. We have set up various instances of cBioPortal for mouse for our clients. For inquiries on setting up cBioPortal for mouse or other services around cBioPortal, feel free to contact us.