Introduction
The development and integration of large language models (LLMs) in the biomedical field has been growing a lot the last three years [1]. LLM’s play a pivotal role in the research of early drug development; they can help predict the mechanism of action, toxicity and other properties of a potentially new drug, all based on literature. However there is a need to have a more transparent way to use an LLM since they are known for their hallucinations. By leveraging annotated databases, hallucinations can be avoided while maintaining transparency. To support researchers in their early drug discovery, The Hyve integrated an LLM into Open Targets Platform (OTP). You can check out this integration on the Open Targets Demo server here.
Approach
BioChatter is used to integrate an LLM into OTP. It is an open source framework which facilitates the use of LLMs in a biomedical context [2]. To avoid hallucinations, a knowledge graph (KG) based on OTP is used to be able to perform retrieval augmented generation (RAG). In other words, The LLM will use the KG to retrieve data and generate an answer. Moreover, a use case question is defined to be able to validate how well the LLM can answer questions based on the KG.
To show the added value of an LLM, the question should be one which cannot easily be answered by just using OTP natively. In OTP, there are disease - gene relationships and gene - GO_term relationships. However, disease - GO_term relationships do not exist. Given the data structures in OTP, the following question is defined:
“Which GO terms are most associated with asthma?”
This question would give insight into which processes or pathways asthma is associated with, identifying new angles to look into for drug discovery research.
Results
Once the KG was modeled and created, the LLM, using GPT-4o, was asked the use case question and answered:
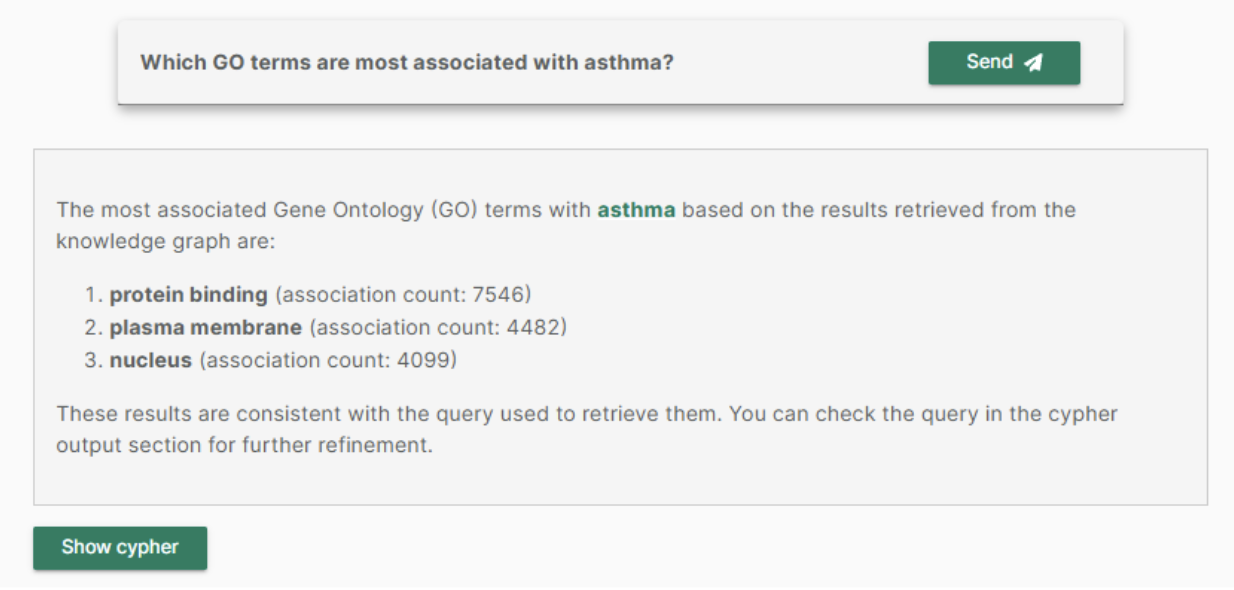
The LLM avoided hallucinations, provided an accurate query, and calculated associations based on the KG, offering transparency. The query can be checked and adjusted if preferred by clicking on the ‘show cypher’ button.
Discussion
The LLM provided only 3 very common GO terms, which might not be very useful. Showing the cypher query and its output will give more insight into which other Go Terms are associated with asthma. From a research perspective a GO term like positive regulation of transcription by RNA polymerase II would be more informative. An increased transcription, or overproduction, is indeed related to inflammatory diseases like asthma. How well the LLM is able to answer questions is highly dependent on the quality of KG and its schema. In this experiment, only a KG prototype was built. Data might be represented in an unnatural way, or uncommon property names are used, which do not serve the LLM in the most optimal way. However, it could still answer the question correctly. It created a semi complicated query, retrieved the result and provided a well constructed answer. The LLM also included the exact query for transparency.
BioChatter demonstrates great potential for drug development research, enabling researchers to uncover insights without requiring expertise in writing database queries like Cypher.
References
[1] Meng, X et al. (2023). The application of large language models in medicine; A scoping review. iScience, Volume 27, Issue 5, doi: 10.1016/j.isci.2024.109713
[2] Lobentanzer, S et al. (2025). A Platform for the Biomedical Application of Large Language Models. nat Biotechnology, doi: 10.1038/s41587-024-02534-3