Janneke
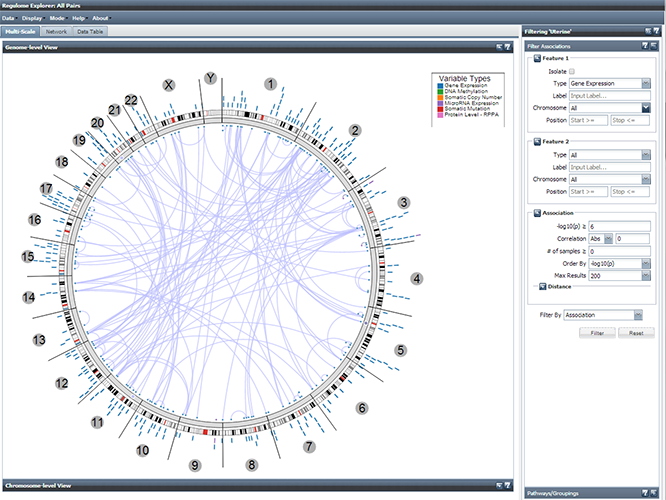
“At this year's NBIC Conference in Lunteren, I was very fortunate to attend a talk by Ilya Smulevich from The Cancer Genome Atlas (TCGA). This was a very inspiring session, where he introduced us to the very rich and heterogenous datasource that has been collected in the area of cancer research. Although we are already working with several datasets from TCGA in our tranSMART projects in the last year, Ilya introduced us to the high-level ideas for collecting this data. Ilya describes this as '...no platform is left behind', meaning that for every patient that data is collected from, they try to collect all datatypes (or leave it out), to make the set as complete as possible. This includes a variety of genomic data, like exome, SNP, methylation, mRNA and miRNA data. A second inspiring part in his talk was the quick tour along the visualisation tools build upon these rich datasets, like the circular genome browser at explorer.cancerregulome.org. Within the TranSMART community the wish for enriched (public) reference datasets in the TranSMART application is expressed quite often, and I think the TCGA repository with it's genomic analytic tools could be of very high value here."
Rianne
“One of the very interesting talks, was the very last talk of the conference. This was a key note lecture by professor Jelle Goeman, titled “Flexible multiple testing”. It addressed the statistical challenges that arise from multiple testing, such as how to control the false positive and false negative rates. This a very relevant, current topic now a days as we are analysing more and more large data sets. Professor Goeman addressed the strengths and limitations of the most commonly used methods to correct for multiple testing; Family Wise Error Rate (FWER) and FDR correction. The first method is very strict and thereby relevant data points may be needlessly excluded. The second method method circumvents this problem, by allowing a certain percentage of false positives among the selected data points (for example among a set of differentially expressed genes). However, if we then were to handpick a certain subset of these data points, for example a set of genes we find particularly interesting, the false positive rate for this subset may differ and may even increase. Thus we lose control of the false positive rate among our subset and we might be drawing conclusions based on false positives. To avoid this problem, while still keeping the false negative rate under control, professor Goeman suggested a solution which is less strict than FWER correction, but does account for handpicking subsets of data after statistical tests were applied. This method involved closed testing and it essentially determines the maximum number of false positives within this set for each possible subset of the data. Therefore, when using this method, the number of false positives is controlled even if a subset of genes is handpicked after applying the statistical test. Note: This method is implemented in the R package called ‘cherry’.”
Jochem
“Many talks on this years NBIC conference were on studies that involve Single Molecule Real Time (SMRT) sequencing on the PacBio platform. This platform allows for very long sequence reads which makes it invaluable in the assembly of genomes with highly repetitive regions. This means we might shed light on currently elusive genomic features. The talk that received the Best Talk Award was on a technique has been developed in the VUmc in Amsterdam, called WISECONDOR. It uses low coverage Next Generation Sequencing to accurately predict fetal genetic disorders from a small percentage of fetal DNA that is present in maternal plasma. Thus it effectively determines chromosomal aberrations, like trisomy 21, without risking miscarriage.”
Jochem was als proud to be active member in the poster presentation, presenting the Integrative Bio-Informatics poster ‘A computational saturated mutation analysis with the MARTINI coarse-grained force field’ by Jochem Bijlard, Jaap Heringa, and K. Anton Feenstra, of the IBIVU Centre for Integrative Bioinformatics, VU University, Amsterdam.
Marcel
“I was lucky to be joining the key note presentations of Marcus Glaesson of the University College of Cork, Ireland regarding (meta)transcriptomics of colonic lesions In Inflammatory Bowel Disease and of Jelle Goeman of Radboud University Nijmegen regarding Flexible Multiple Testing. I also attended the parallel session regarding infrastructure, data and technology and I have met up with the RSG network of young Bioinformaticians. I liked it that I could enjoy such a wide variety of topics in such a short time. What I really liked was the fact that I could meet up with students, PHDs and professionals of Bioinformatica companies to hear what’s driven them and their work.“
Seth
“NBIC a conference to remember. Although I am not a biologist at all, the given presentations were still interesting. A few topics that crossed the board and caught my attention are: PPI (proteint protein interaction), PTM (post translational modificication) and Asexual reproduction. The impresseion I got from the conference is positive overall, due to the clear indepth presentations and presenters professionalism.”
See also explorer.cancerregulome.org or http://cancergenome.nih.gov/abouttcga/overview. More general info and downloads can be found on the NBIC site.