Compare your samples based on cancer gene signatures
Anyone who works with transcriptomics data knows about Gene Set Enrichment Analysis — a powerful method to compare the behaviour of gene sets (aka gene signatures, e.g., pathways) in different sample groups. But have you ever been in a situation where you need to compare samples to each other based on their expression profile? Maybe when you want to find biological processes that are differentially active across the phenotype of interest? Or if you found a correlation between gene signatures and response to drug X and want to predict the response of new patients based on their gene signatures? Maybe, as a researcher you are interested to know which pathways are enriched/depleted in a certain (sub)type of cancer? Or, as an oncologist, you are looking for correlations between mutations/alterations in specific genes and increased or decreased activity in a specific pathway?
This is where Gene Set Variation Analysis (GSVA) comes into play. GSVA is a statistical method that allows you to detect consistent activity in pathways and other signatures over a group(s) of samples. In other words, GSVA is a sample-wise enrichment analysis method, which provides great power to detect even subtle pathway activity changes over a sample population.
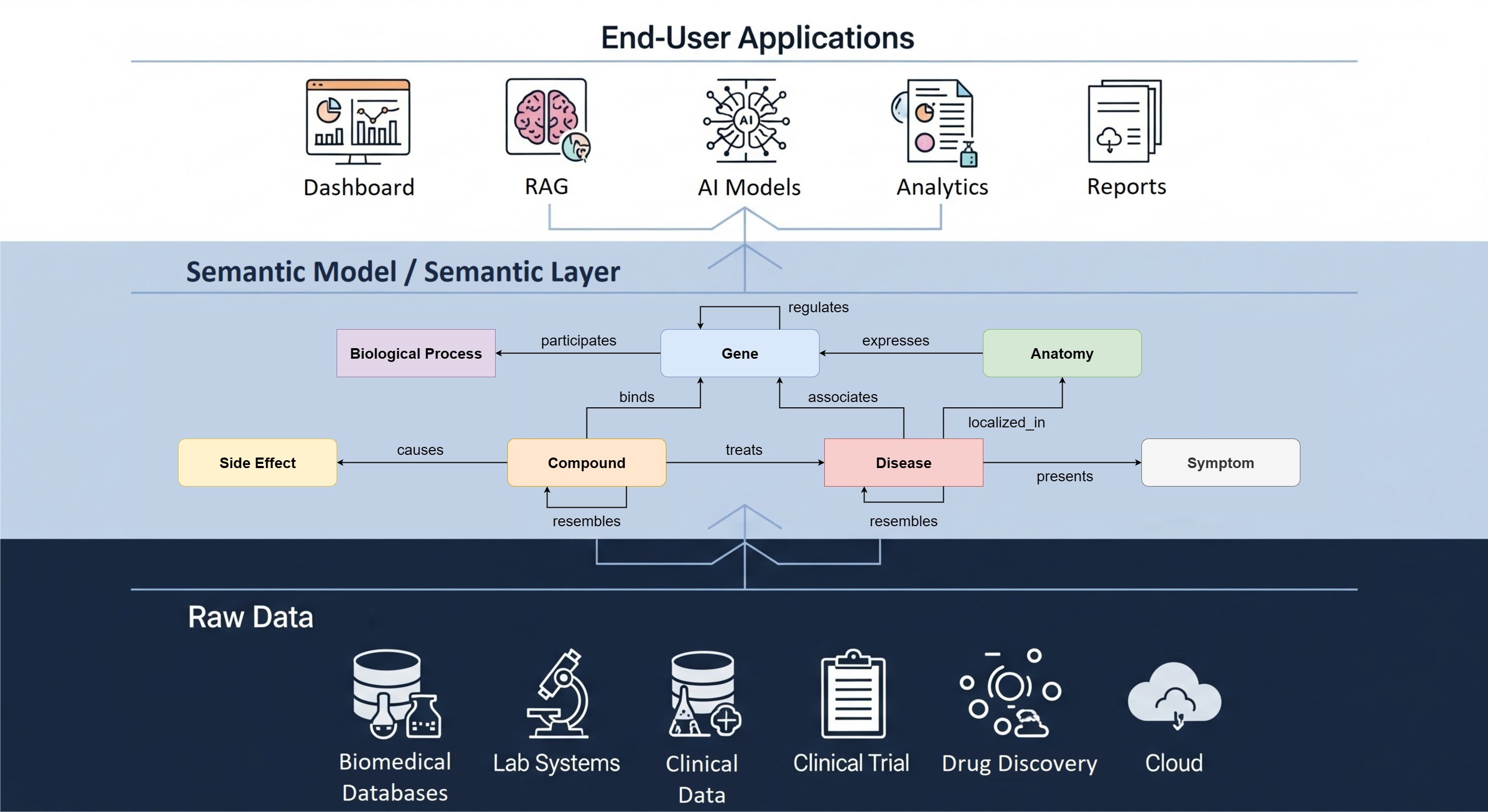
The Hyve has recently implemented GSVA functionality into cBioPortal, a popular open source cancer genomics visualization and analysis platform. To look at your GSVA scores and integrate them with the rest of the data, you don’t have to download expression and mutation profiles of hundreds of cancer samples and develop tools to analyze everything together anymore. Using the functionality of cBioPortal, you can now easily visualize your GSVA results and do further integrated analysis, e.g., look at the relationship between your gene signatures and clinical data or genetic alterations.
GSVA visualisation

We implemented the support for gene sets and pre-calculated GSVA scores for the gene sets: now in cBioPortal you can visualize your GSVA scores per sample, cluster them hierarchically and analyze their relationship with some molecular features of your samples (scroll down to see a few snapshots of this new cBioPortal functionality). GSVA scores represent the degree to which the genes in the gene set are coordinately up- or downregulated within that sample. The new functionality includes the support for the hierarchical clustering of heatmaps in the oncoprint of cBioPortal, which is a general purpose feature we also contributed to the main cBioPortal code base during the development of this project (figure below shows it also being used for regular gene based expression data).

Use the power of cBioPortal to its fullest and run integrated analyses of GSVA scores of selected gene signatures and gene expression, mutation and copy number alteration profiles of your samples.

cBioPortal provides you with the table of gene sets that are highly correlated with a gene or gene set of your choice

In summary, GSVA support enables a new dimension of exploratory analysis in cBioPortal, allowing researchers to search and find patterns at the level of molecular processes (i.e. multiple genes that are known to work in concert), instead of exploring the data at the level of individual genes.
If you want more information on GSVA and its implementation in cBioPortal, or if you need help with loading your GSVA data in cBioPortal please contact us.