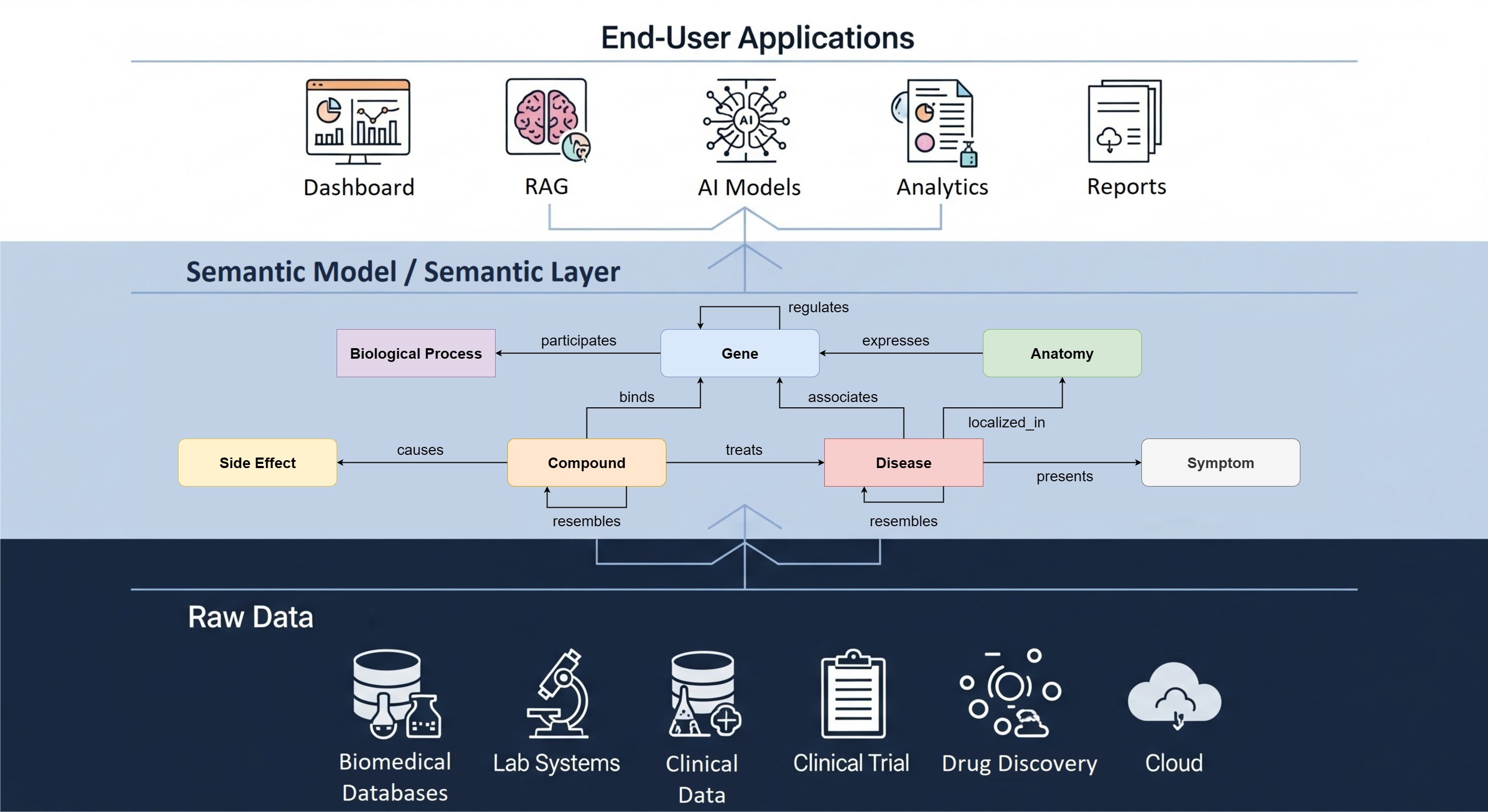
The OncoPrint view in cBioPortal is a user friendly visualization for genetic alterations. All types of data from a cancer genomic study are integrated in the OncoPrint view: in one overview you can see mutations, expression levels and copy number alterations for the queried genes. These genetic alterations can also be associated with clinical attributes like the overall survival status of a patient, and heatmaps with z-score values from the different analysis types can be added.

At The Hyve, we developed the functionality to download the OncoPrint data in tabular format. This functionality will generate a single file with a column for each sample and a row for each gene alteration. This feature is included in the public release of cBioPortal (GitHub) since version 1.9.0.
The file can be directly imported in e.g., R or Excel to do further downstream analysis of the selected data subset and genetic alterations. This makes it very easy to combine analyses from the different data types.

So you can for example create the following histogram in cBioPortal:

To recreate this Histogram, use the following code:
# Import necessary libraries
library(ggplot2)
library(scales)
library(reshape2)
# Downloaded oncoprint data from Breast Cancer TCGA provisional,
# selecting all genomic profiles and querying BRCA1 and BRCA2
mutations = read.csv("~/Downloads/PATIENT_DATA_oncoprint.tsv", sep="\t", skip=2, nrow=2, row.names=1)
# Combine mutation types for both genes in one row
combined_mutations = melt(t(mutations[,2:ncol(mutations)]))
# Set more informative columnnames
colnames(combined_mutations) = c("var", "gene", "mutation_type")
# Add numeric class definition for mutation type
combined_mutations = data.frame(mutation_group=as.numeric(combined_mutations$mutation_type), combined_mutations)
# Remove rows where mutation_type is NA or empty
combined_mutations = combined_mutations[!combined_mutations$mutation_type %in% c('',NA),]
# Create barchart with mutation types in genes BRCA1 and TP53
ggplot(combined_mutations, aes(x=gene, y=mutation_group, fill=mutation_type)) +
labs(x="", y="", title="Mutation types in BRCA1 and TP53", fill="Mutation Type") +
scale_fill_manual(breaks = c("Inframe Mutation (putative passenger)", "Inframe Mutation (putative driver)", "Missense Mutation (putative driver)", "Missense Mutation (putative passenger)", "Truncating mutation (putative driver)"),
values=c("yellow2", "green3", "turquoise3", "deeppink2", "navy")) +
scale_x_discrete(breaks=c("BRCA1", "TP53"), labels=c(paste0("BRCA1\n", length(combined_mutations$gene[combined_mutations$gene == "BRCA1"]), " mutations"), paste0("TP53\n", length(combined_mutations$gene[combined_mutations$gene == "TP53"]), " mutations"))) +
scale_y_continuous(labels=percent_format()) +
geom_bar(position = "fill", stat="identity")
If you need any support, please contact us.