Introduction to Open Targets Genetics
Genome-Wide Associations Studies (GWAS) have become a widely adopted resource in the field of preclinical drug discovery. Genetic variants found in GWAS studies that are associated with genes have become an important factor in the drug discovery pipeline. A GWAS study however can have thousands of potential targets, so pinpointing the right causal variants is important. Our Open Targets Genetics website helps with the difficult task of deciding which genes are involved in a phenotype. Using many different data sources, a score is calculated that associates each variant to one or more genes. With colocalization information, causal variants can be found for a disease area of interest. For more information about Open Targets Genetics, please read our previous blog post.
The data processing pipeline
Open Targets Genetics contains GWAS studies from multiple large data sources such as the UK Biobank, Finngen, and the GWAS catalog. To analyze a GWAS study properly, different methods have been developed to, for example, extract variants (fine-mapping), compare against other studies (colocalization) and assign genes to variants (v2g, read: variant-to-gene). Open Targets Genetics incorporates these methods into a pipeline that extracts valuable information from raw GWAS studies. The resulting data is stored in the Open Targets Genetics Portal, where you can analyze or retrieve it.
The web application
The Open Targets Genetics web application contains many features to analyze and retrieve GWAS-related data. The application was developed with the aim of optimal user experience in mind. The search box brings you to any of the three main pages: the study, gene, or variant page. Each page contains elements that provide all available information on a gene, a variant, or a study. For example, the study page contains a Manhattan plot with an overview of all interesting variants derived from the corresponding GWAS study. The variant page contains a PheWAS plot that shows all the studies that include that variant. The gene page provides information on the loci which functionally implicate a gene and links out to detailed information on the gene and drugs targeting it.
The Hyve's Open Targets Genetics instance
Even though Open Targets Genetics comes with many valuable components to identify potential genes through GWAS, there is always room for improvement. The field of human genetics is a very active research area in which new methods are constantly being developed. It’s impossible to integrate all of them into one application. However, we can imagine that you have some preferred methods or you have developed a new one yourself. In this case, we could add these and extend the Open Targets Genetics application. This would further enrich the wealth of information already available on the website. Recently, we have implemented a number of popular methods and visualizations in our new Open Targets Genetics installation, which we will describe in the following sections.
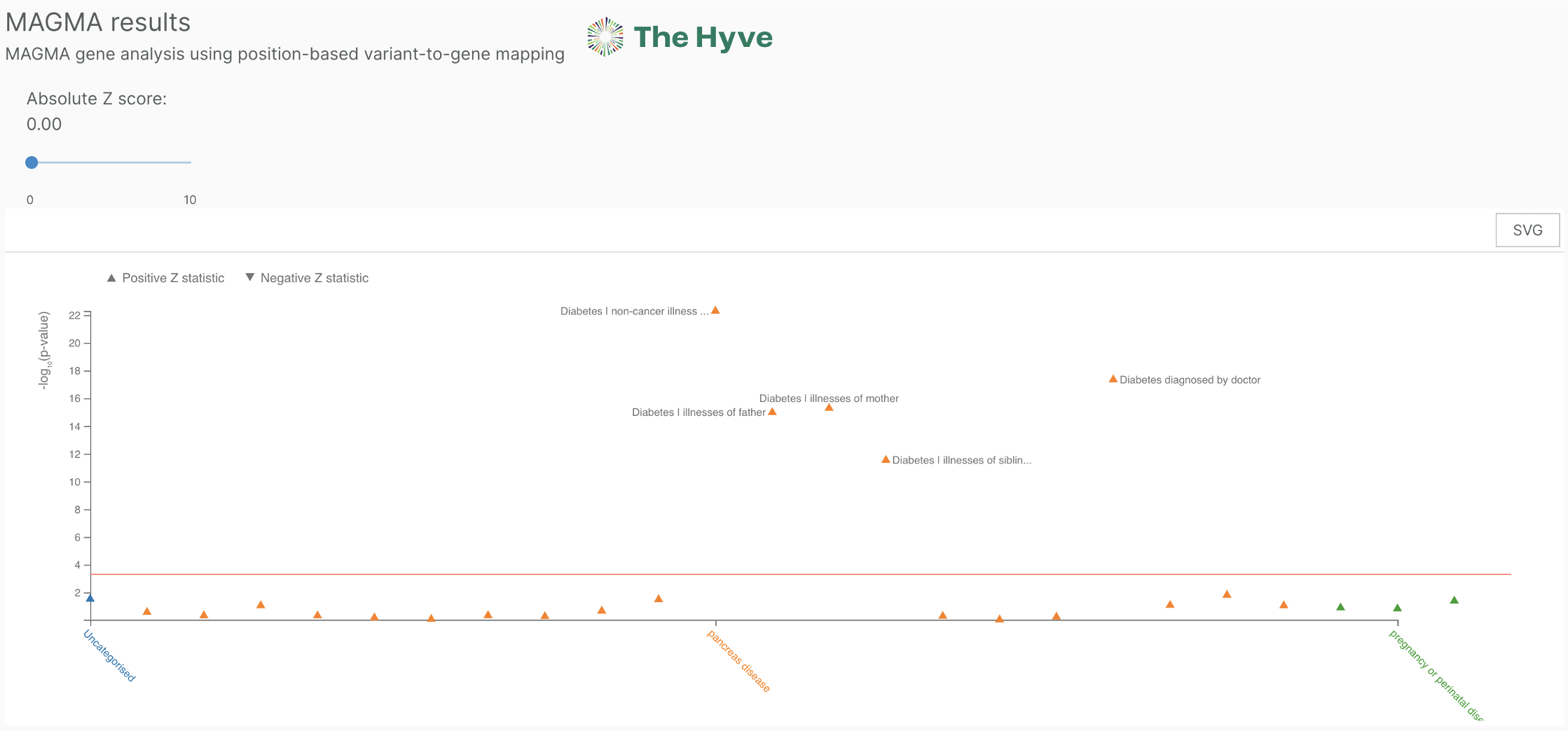
MAGMA
MAGMA is a method developed for gene or gene-set analysis based on a multi-regression model. You can find the MAGMA results on our study and gene page. On the gene page, all studies are shown that are linked to the gene via MAGMA. The section contains a slider where you can filter results by setting a cutoff for the z-score.
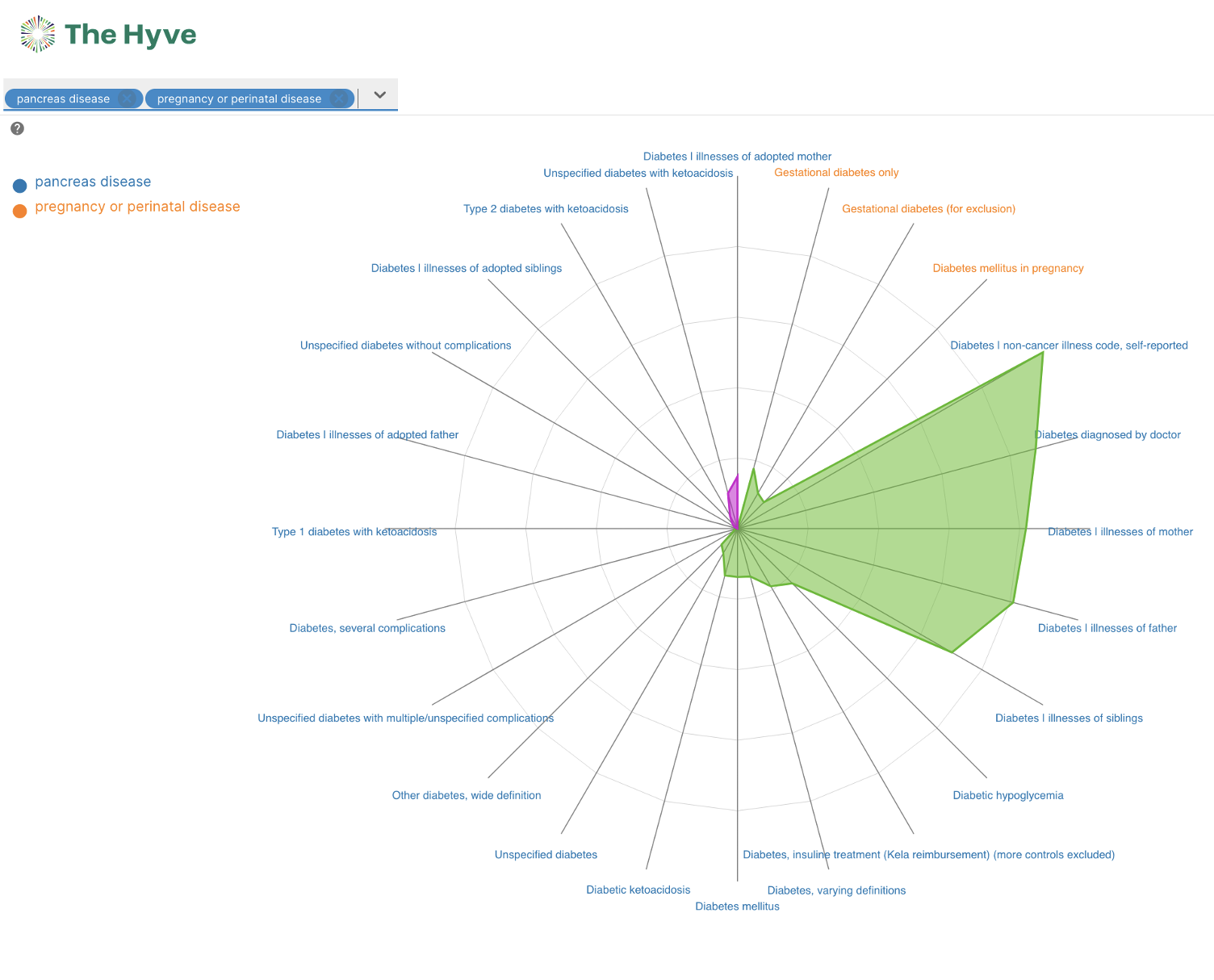
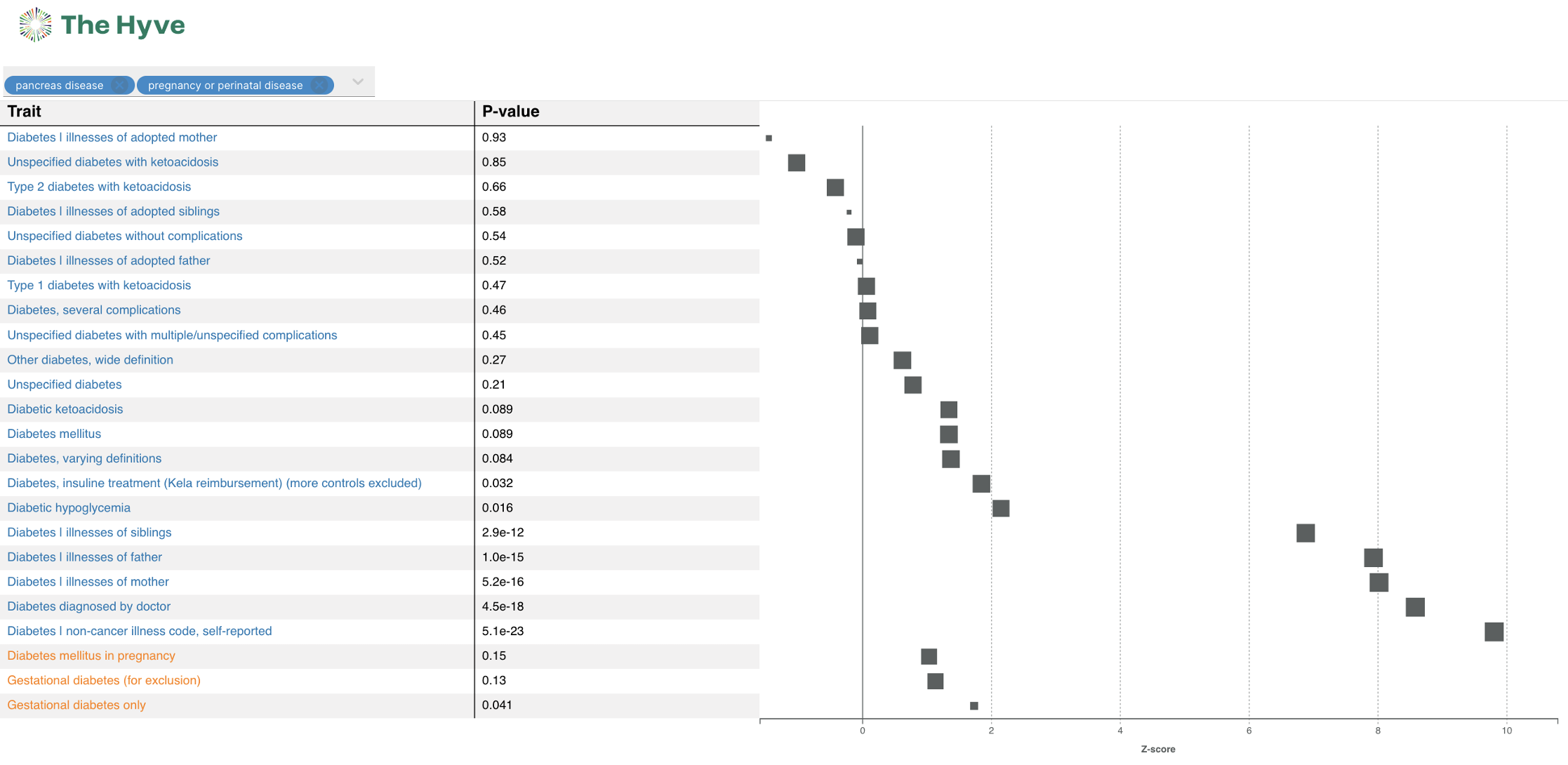
Radar and Forest plots
A radar and forest plot have been added to the variant and gene page. Both these plots make it possible to compare different groups of phenotypes, called trait categories. By comparing these groups, you might find new insights or outliers relevant to your research.
Want your own customized Open Targets Genetics instance?
Hopefully, this blog post has made you as excited about the customizability of Open Targets Genetics as we are. The changes we have made in the past months are just some examples of what is possible. Don’t be afraid to think out of the box. If you have any questions regarding Open Targets Genetics and the possibilities that it might bring to your research, feel free to contact us.
At The Hyve, we’re always on the lookout for new features to maintain and extend our Open Targets instance So if you want to learn about those, don’t forget to check out our website from time to time.